Elucidating natural microbial communities
We use microfluidic tools to deconstruct and systematically study complex microbial communities from natural environments such as the human body (gut and vagina) and ecosystems (Lake Erie).
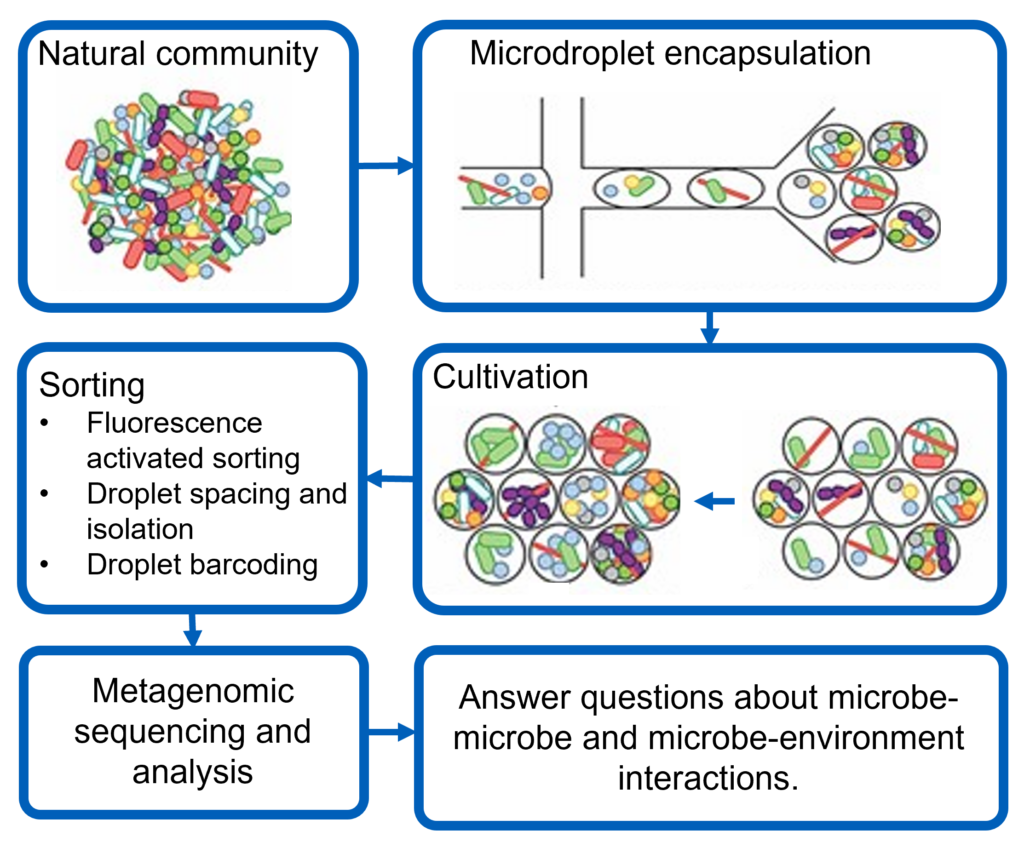
Projects:
- Microfluidic-based investigation of autotroph-heterotroph interactions in Lake Erie cyanobacterial bloom aggregates
- Quantitative meta-transcriptomic platform for microdroplet encapsulated microbial consortia
- Microfluidic elucidation of microbial consortia capable of complex carbohydrate utilization in the human gut (past)
- Elucidating Bacterial Interactions in the Vaginal Microbiome Using Microdroplet Co-cultivation Technology (past)
Ongoing projects
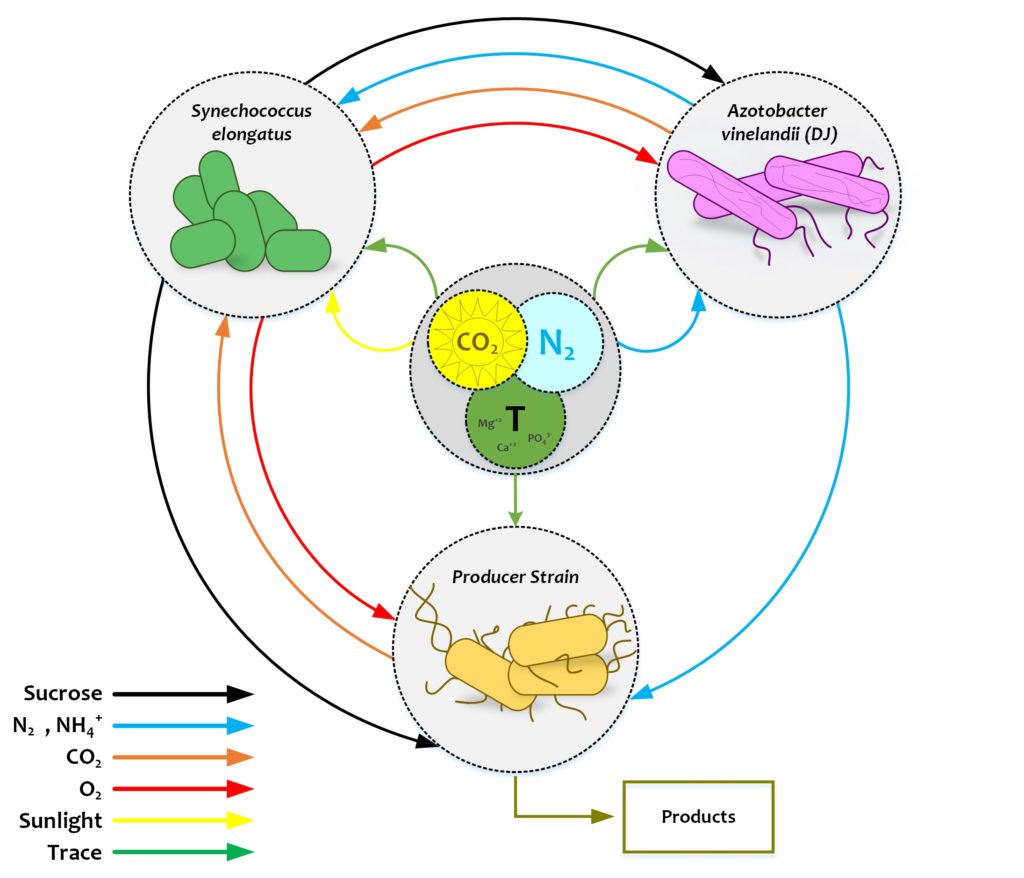
Developing, understanding, and harnessing modular carbon/nitrogen-fixing tripartite microbial consortia for versatile production of biofuel and platform chemicals.
Current students: Sofía Camarero
Past students: David Carruthers, Andrew Kim
Collaborators: Prof. Maciek Antoniewicz, Prof. Andrew Allman, Prof. Sujit Datta (Princeton Chemical Engineering)
Current Funding: Department of Energy
Engineering of synthetic microbial consortia has emerged as a new and powerful biotechnology platform. To date, most microbial consortia have focused on biofuel development, though they also have enormous potential in the production of biobased commodity chemicals. Our work describes a tripartite system in which three microbes with different specializations can convert sunlight, carbon dioxide, and atmospheric nitrogen into chemical precursors for the production of biofuels, biopolymers such as polyhydroxyalkanoates (PHAs) and polylactic acid (PLA), and other products without energetically or monetarily expensive nutrient inputs. This system is an attractive, sustainable alternative to fossil fuel analogues. Specifically, Azotobacter vinelandii, a nitrogen-fixing bacterium that secretes ammonia, and Synechococcus elongatus, a photosynthetic cyanobacterium that secretes sucrose, form a symbiotic chassis hypothesized to support a third producer strain. Escherichia coli and Corynebacterium glutamicum are two archetypal bacterial species previously modified to produce an enormous array of chemicals. Given that both species can naturally grow on ammonia and sucrose, this will facilitate a tailored three-part system with drop-in target production.
Read David Carruthers’ (PhD’20) Thesis: Engineering Modular Synthetic Microbial Consortia for Sustainable Bioproduction From CO2
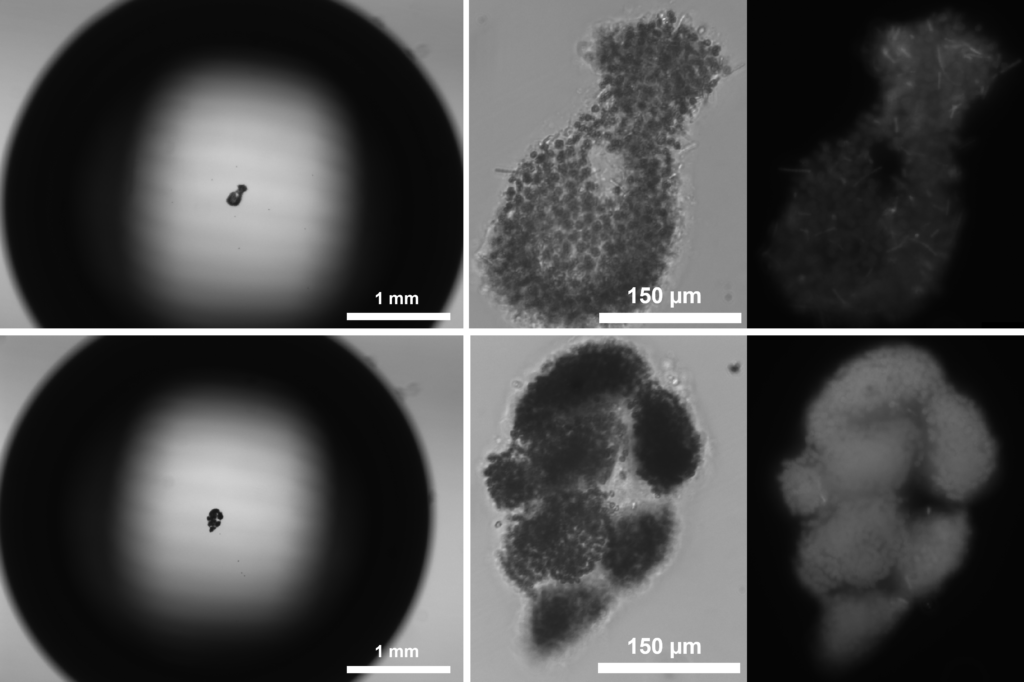
Microfluidic-based investigation of autotroph-heterotroph interactions in Lake Erie cyanobacterial bloom aggregates
Current students: James Tan
Collaborator: Prof. Gregory Dick, Department of Earth and Environmental Sciences
Lake Erie, reoccurring harmful cyanobacterium blooms are a threat to water resources and ecosystem health. Microcystis aeruginosa, the cyanobacterial producer of harmful toxins, tends to form micron-scale colonies in association with other heterotrophic bacteria, the identity and function of which are not known. With the use of microfluidic technology to study these colonies with high-resolution, we hope to understand the identity, metabolic interactions, and ecosystem function of these colonies and their impact on toxin production.
Quantitative meta-transcriptomic platform for microdroplet encapsulated microbial consortia
Current students: James Tan
Although metagenomics of microbial cells or populations has enabled the recovery of genomes from rare and phylogenetically novel bacteria from single microdroplets, there lacks a toolset to perform transcriptomics on individual bacterial populations to determine molecular interactions. As such, we are developing a microfluidic and molecular biology framework to develop this and elucidate novel metabolic interactions in microbial communities.
Past Projects
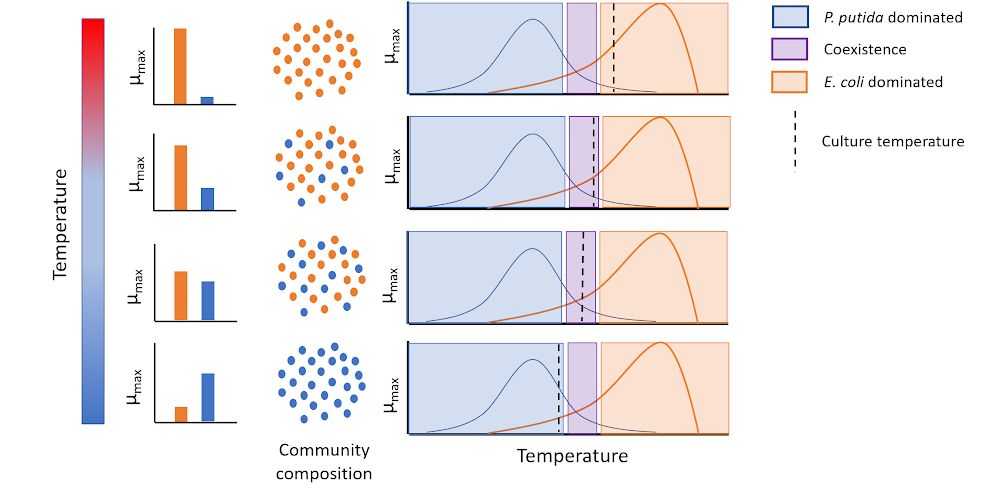
Temperature Cycling to Control Population Dynamics (Adam Krieger, PhD’20)
We are combining ecological theory, mathematical modeling, and synthetic biology approaches to determine how community population dynamics respond to constant or cycling changes in temperature. A deeper understanding of the mechanisms involved in this response makes it possible to use temperature regulation to enable coexistence of communities and measure their composition as it changes. We hope to develop a mathematical model that will allow us to reliably predict whether multiple species will be able to coexist at a given temperature.
Tunable chromosomal integration of sucrose catabolism in E. coli K12 (David Carruthers, PhD’20)
Sucrose is the most abundant naturally occuring disaccharide. Most industrially biofermentation plants use feedstocks of fructose and glucose, which are derived by hydrolyzing sucrose from sugar cane or beets, whcih pose implciations for cost and energty targets associated with substrate production. E. coli is a cononical organism to study metabolism and fundamental to molecular biology. Recent advances have illustrated that chromosomal positioning can dramatically influence gene expression. This work explores how varied positions can impact sucrose catabolism and hence growth rates.
Microfluidic elucidation of microbial consortia capable of complex carbohydrate utilization in the human gut (James Tan)
The utilization of complex carbohydrates from dietary fiber by human colonic bacteria has been shown to be a major factor in maintaining host health, particularly through the production of short-chain fatty acids, such as acetate, butyrate, and propionate. This function is performed in multiple trophic steps by consortia, the identity of which is not exactly known and can be highly variable from individual to individual. Microfluidic technology has the capability to encapsulate subsets of microbial populations in well-localized compartments for a high-throughput and high-resolution study of these consortia and their utilization of complex carbohydrates.
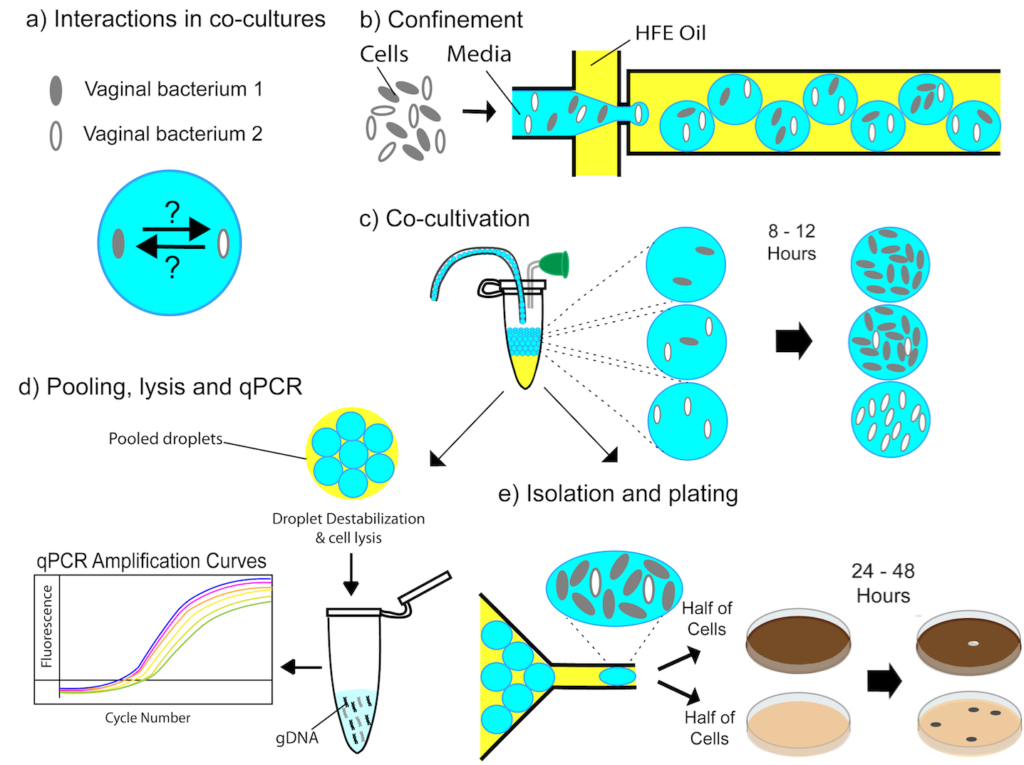
Elucidating Bacterial Interactions in the Vaginal Microbiome Using Microdroplet Co-cultivation Technology (Corine Jackman PhD’19)
Overgrowth of strict anaerobic bacteria in the vagina can lead to bacterial vaginosis (BV), the most common vaginal disorder among women of reproductive age and is also responsible for millions of health care visits per year. Homeostasis of the human vaginal microbiome (HVM) is significant for women’s health. However, the ecological roles of many vaginal species remain unclear and current approaches for investigating them have severe limitations. A major focus of this research is to understand the roles of individual species in microbial communities. To develop more insight into these interactions, we have employed microdroplets to co-cultivate vaginal bacteria. Past work has served as a conceptual and technological framework to characterize a well-established inter-species interaction between two vaginal bacterium in microdroplets. Ongoing work includes encapsulating vaginal bacterium with specimen from the host to explore their effects on inter-species interactions.